Variant Impact Predictor database (VIPdb), version 2.01
407 variant impact predictors
Lin YJ, Menon AS, Hu Z, Brenner SE. Variant Impact Predictor database (VIPdb), version 2: trends from three decades of genetic variant impact predictors. Hum Genomics (2024) 18(1):90. DOI
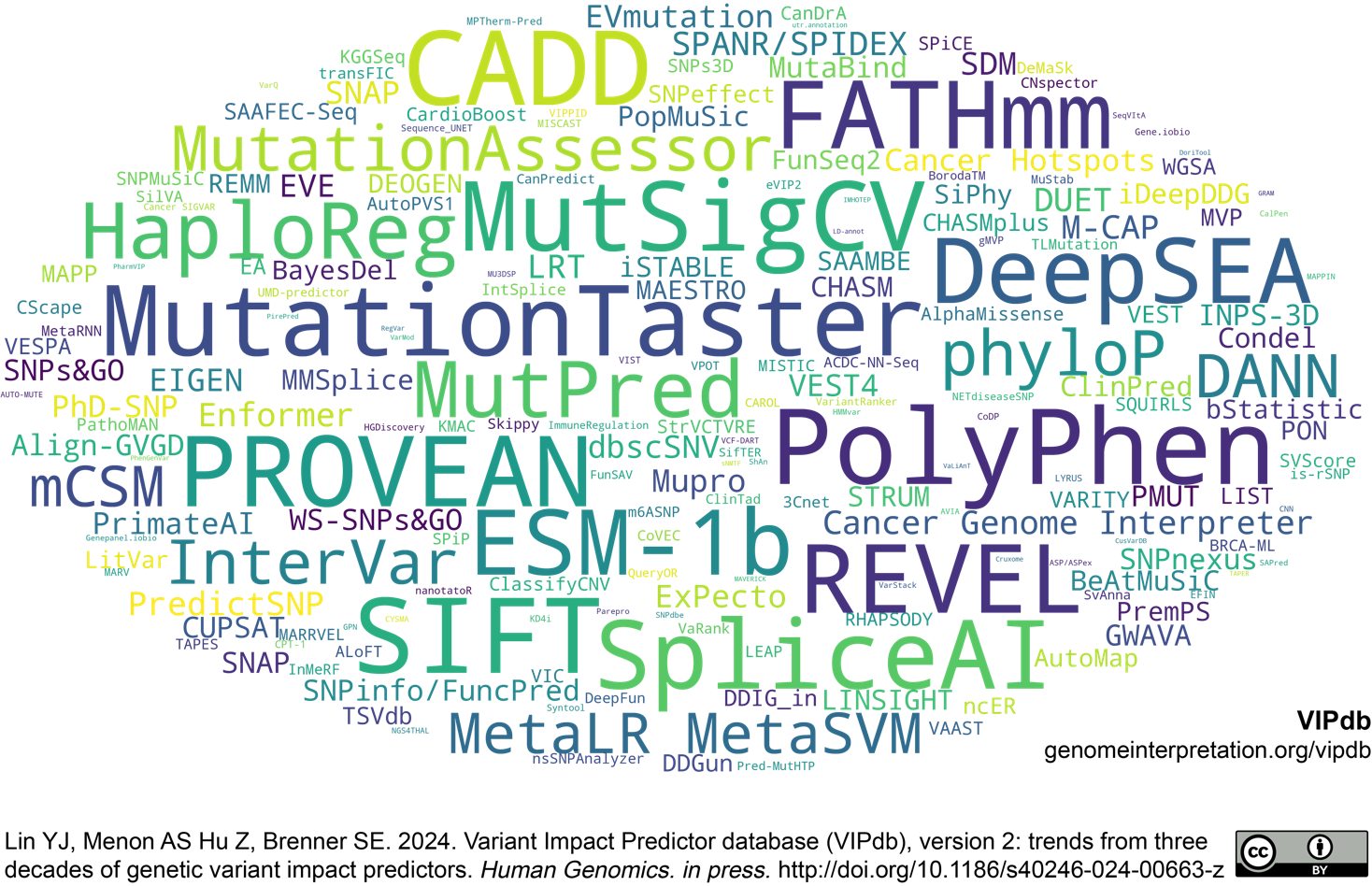
278 core VIPs scaled by log of citations 2021 to 2023. Download vector PDF, PNG, or high-resolution PNG.
Variant interpretation is essential for identifying patients’ disease-causing genetic variants amongst the millions detected in their genomes. Hundreds of Variant Impact Predictors (VIPs), also known as Variant Effect Predictors (VEPs), have been developed for this purpose, with a variety of methodologies and goals. To facilitate the exploration of available VIP options, we have created the Variant Impact Predictor database (VIPdb).
The Variant Impact Predictor database (VIPdb) version 2.01 presents an updated collection of 407 VIPs developed over the past three decades, summarizing their characteristics, such as publication details, access information, and citation patterns.
VIPdb version 2 adds data to inform clinical decision-making. We incorporated calibrated threshold scores recommended by ClinGen for clinical use (Pejaver et al., 2022) with ACMG/AMP guidelines for variant classification (Richards et al., 2015). Additionally, we included community assessment results from the CAGI6 Annotate All Missense / Missense Marathon challenge (Rastogi et al., 2024) to enable users to compare method performance.

Citation trend of 278 core VIPs (1998 to 2023). Font sizes correspond to the logarithm of citation counts for each period, and word cloud heights are scaled by the logarithm of the annual citation averages. The top 10 most cited core VIPs during the period are listed. (Core VIPs are those methods primarily designed for variant impact prediction and are not classified as databases)
VIPdb version 2.01
VIPdb version 2.01 database spreadsheet can be downloaded here.
VIPdb version 2.01 description can be downloaded here.
VIPdb version 2.01 word cloud for 2021 to 2023 can be downloaded in vector PDF, PNG, or high-resolution PNG format.
VIPdb version 2.01 word cloud time course figure can be downloaded in vector PDF, PNG, or high-resolution PNG format.
VIPdb version 1
VIPdb version 1 (Hu et al., 2019) database spreadsheet can be downloaded here.
Release history
VIPdb version 2.01: Aug 26, 2024
VIPdb version 2.00: July 10, 2024
VIPdb version 1: July 8, 2019
Funding
NIH NHGRI/NCI CAGI grant U24 HG007346
UCB-Taiwan Fellowship
Contact
Steven E. Brenner: brenner@compbio.berkeley.edu
Yu-Jen (Jennifer) Lin: jenniferyjlin@berkeley.edu
References
Hu Z, et al. VIPdb, a genetic Variant Impact Predictor database. Hum Mutat (2019) 40(9):1202-1214. PubMed
Lin YJ, et al. Variant Impact Predictor database (VIPdb), version 2: trends from three decades of genetic variant impact predictors. Hum Genomics (2024) 18(1):90. PubMed
Pejaver V, et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am J Hum Genet (2022) 109(12):2163-2177. PubMed
Rastogi R, et al. Critical assessment of missense variant effect predictors on disease-relevant variant data. Hum Genet (2025) 144(2-3):281-293. PubMed
Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17(5):405-424. PubMed
Last updated: December 27, 2025
