Stability and catalytic efficiency of mitogen-activated protein kinase 3
Challenge: MAPK3
Variant data: registered users only
Last updated: 12 October 2021
This challenge is closed.
How to participate in CAGI6? Download data & submit predictions on Synapse
Make sure you understand our Data Use Agreement and Anonymity Policy
Summary
MAPK3 (ERK1) is active as serine/threonine kinase in the Ras-Raf-MEK-ERK signal transduction cascade that regulates cell proliferation, transcription, differentiation, and cell cycle progression. MAPK3 is activated by phosphorylation which occurs with strict specificity by MEK1/2 on Thr202 and Tyr204, and may also act as a transcriptional repressor independent of its kinase activity. A library of twelve missense variants selected from the COSMIC database was assessed by near and far-UV circular dichroism and intrinsic fluorescence spectra to determine thermodynamic stability at different concentrations of denaturant. These data were used to calculate a ΔΔGH20 value; i.e., the difference in unfolding free energy ΔGH20 between each variant and the wildtype protein, both in phosphorylated and unphosphorylated forms. The challenge is to predict these two ΔΔGH20 values and the catalytic efficiency (kcat/km)mut/(kcat/km)wt, as determined by a fluorescence assay, of the phosphorylated form for each MAPK3 variant.
Background
Human MAP kinases (Mitogen-activated protein kinases), also known as extracellular signal-regulated kinases (ERKs), are proteins involved in the integration of multiple biochemical signals and in several cellular processes ranging from transcription regulation and development, to differentiation and proliferation (Lavoie et al., 2020). MAPK3 is activated through its phosphorylation by upstream kinases (Roskoski, 2012). Upon activation, MAPK3 translocates to the nucleus to phosphorylate specific nuclear targets. Independently of its kinase activity, MAPK3 may act as a transcriptional repressor and has been identified as a moonlighting protein based on these multiple distinct functions (Lu et al., 2018).
These functions may vary depending on the cellular context (Raman et al., 2007) and include cell growth, adhesion, survival and differentiation through transcriptional regulation, translation, and cytoskeletal rearrangements. Around 160 putative protein substrates have been described for MAP kinases. The substrates localized in the nucleus seem to be responsible for the stimulation and regulation of transcription, while the substrates localized in cytosol and organelles are responsible for translation, mitosis and apoptosis and may take part in the checkpoint of spindle assembly. Owing to their biological importance, protein kinases represent an important target of biomedical research and of a large part of drug discovery research (Moret et al., 2020 ).
The single amino acid variants included in the MAPK3 challenge were selected from the hundreds of variants with high mutation rates detected in tumor samples reported in the COSMIC (Catalog of Somatic Mutations in Cancer) database. These are somatic variants distributed along the protein sequence which could additionally be expressed as a soluble recombinant protein in E. coli.
Experiment
Unphosphorylated human MAPK3 (ERK1) variants were obtained with specific mutagenesis primers by PCR, using wildtype protein as a template. Wildtype proteins and mutants are then expressed in E. coli cells Rosetta with phosphatase and purified by affinity chromatography by a 2‐ml prepacked His trap column (GE Healthcare, Chicago, IL).
The phosphorylated (active) form of MAPK3 wildtype and variants were obtained by using the plasmid pGEX-KG-MEKR4F (the active mutant of MEK1) plasmid (kindly provided by Professor M. Cobb from Southwestern University, TX, USA) for the co-expression of the wildtype and the mutants. MAPK3 wildtype and variants were expressed as N-terminally His-tagged proteins in E. coli cells BL21(DE3) and purified by ion exchange and affinity chromatography by a 2‐ml prepacked His trap column (GE Healthcare, Chicago, IL) .
This protocol introduces heterogeneity in the protein mixture, with up to one or two more phosphorylation sites being potentially generated, in addition to the two canonical sites (Thr202, Tyr204). To check the MAPK3 phosphorylation, we performed western blot analysis with the Antibody cat. #44-680G in IHC (Invitrogen).
The thermodynamic stability was measured at increasing denaturant concentration by monitoring the spectral changes (far-UV circular dichroism and intrinsic fluorescence emission) induced by the denaturant. The variation of unfolding free energy (ΔG) is calculated fitting the spectral changes at zero denaturant concentration (Walters et al., 2009) (ΔGH2O). The data are the average of at least three replicates on different purification batches.
The catalytic activity of the MAPK3 wild-type and variants is determined by a fluorescence assay as described in Shults et al. (2003, 2005, 2006). This method is based on Chelation-Enhanced Fluorescence (ChEF) upon substrate peptide phosphorylation by the kinase.
Prediction challenge
For each variant, participants are asked to predict the ΔΔGH20 value for the phosphorylated and unphosphorylated forms. The ΔΔGH20 is the difference in unfolding free energy (ΔGH20) between the mutant and wildtype proteins, in kcal/mol. Furthermore, the prediction of the ratio between the catalytic efficiency [rkcat/km = (kcat/km)mut/(kcat/km)wt] of the mutant and the wildtype.
The experimental free energy change values (ΔGH20) of the wildtype protein in phosphorylated and unphosphorylated forms fitted with a two and three-states curves are the following:
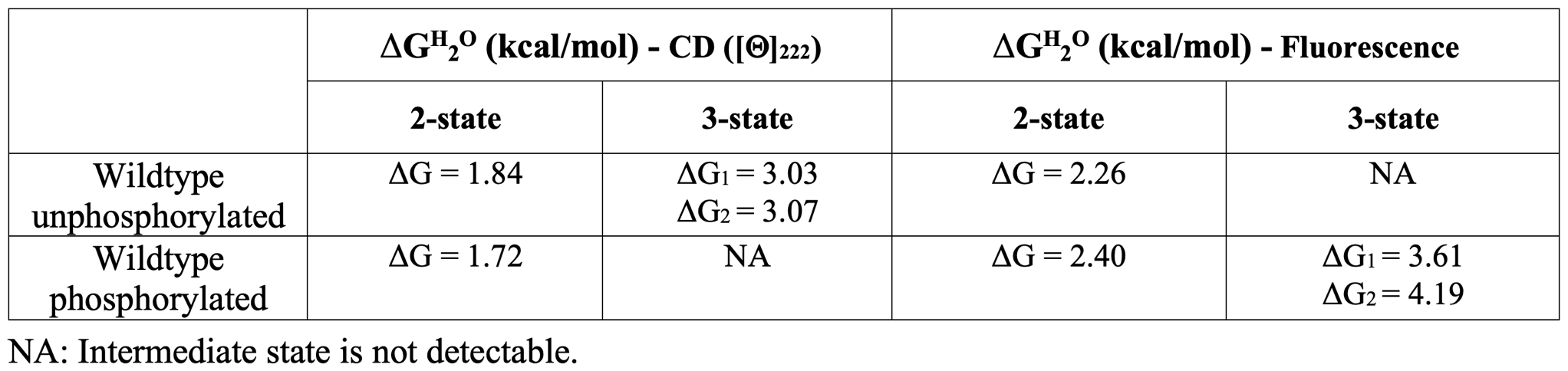
The experimental efficiency of the catalytic activity (kcat/km) of the wildtype MAPK3 in the phosphorylated form is 0.65 s-1 μM-1.
Prediction submission format
The prediction submission is a tab-delimited text file. The organizers provide a template file, which must be used for submission. In addition, a validation script is provided, and predictors must check the correctness of the format before submitting their predictions. Each data row in the submitted file must include the following columns:
- AA substitution: Single-letter amino acid substitution and position; e.g., relative to the UniProt protein P27361 (MK03_HUMAN).
- Value: ΔΔGH20 (unphosphorylated)
- Standard deviation: SD of the prediction in column 2
- Value: ΔΔGH20 (phosphorylated)
- Standard deviation: SD of the prediction in column 4
- Value: rkcat/km
- Standard deviation: SD of the prediction in column 6
- Comment: optional brief comment on the basis of the predictions
In the template file, cells in columns 2-8 are marked with a "*". Submit your predictions by replacing the "*" with your value and note that you can choose to predict free energy but not catalytic activity or vice versa. No empty cells are allowed in the submission. For a given subset, you must submit predictions and standard deviations for all or none of the variants; if you are not confident in a prediction for a variant, enter an appropriate large standard deviation for the prediction. Optionally, enter a brief comment based on the prediction. If you do not enter a comment on a prediction, leave the "*" in those cells. Please make sure you follow the submission guidelines strictly.
In addition, your submission should include a detailed description of the method used to make the predictions, like the style of the Methods section in a scientific article. This information must be submitted as a separate file.
File naming
CAGI allows submission of six models per team, of which model 1 is considered to be primary. You can upload predictions for each model multiple times; the last submission before deadline will be evaluated for each model.
Use the following format for your submissions: <teamname>_model_(1|2|3|4|5|6).(tsv|txt)
To include a description for your method(s), use the following filename: <teamname>_desc.*
Example: if your team’s name is “bestincagi” and you are submitting predictions for your model number 3, your filename should be bestincagi_model_3.txt.
Additional information
More information on human MAPK3 is reported in UniProt accessible with the identifier P27361 (MK03_HUMAN). Many structures of human MAPK3 have been determined by X-ray crystallography. Among them in the PDB are available with codes 4QTB and 2ZOQ. 4QTB is a structure in complex with a small molecule inhibitor mapping from position 24 to 374. 2ZOQ is a structure with phosphorylated residue in position 204 mapping from position 24 to 375.
Assessment
This challenge will be assessed by Emidio Capriotti, University of Bologna, Italy.
Related challenges
Data provided by
The dataset is provided by the group of Roberta Chiaraluce and Valerio Consalvi, Sapienza University, Rome, Italy.


References
Lavoie H, et al. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol (2020) 21: 607-632. PubMed
Lu Z, Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci (2018) 43(4), 301-310. PubMed
Moret N, et al. A resource for exploring the understudied human kinome for research and therapeutic opportunities. (2020) bioRxiv 2020.04.02.022277. bioRxiv
Motta M, et al. Enhanced MAPK1 function causes a neurodevelopmental disorder within the RASopathy clinical spectrum. Am J Hum Genet (2020) 107(3): 499-513. PubMed
Raman M, et al. Differential regulation and properties of MAPKs. Oncogene (2007) 26: 3100-12. PubMed
Roskoski R Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res (2012) 66: 105-143. PubMed
Shults MD, Imperiali B. Versatile fluorescence probes of protein kinase activity. J Am Chem Soc (2003) 125(47): 14248-14249. PubMed
Shults MD, et al. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat Methods (2005) 2(4): 277-283. PubMed
Shults MD, et al. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal Biochem (2006) 352(2): 198-207. PubMed
Walters J, et al. Practical approaches to protein folding and assembly: spectroscopic strategies in thermodynamics and kinetics. Methods Enzymol (2009) 455: 1-39. PubMed
Revision history
04 August 2021: initial announcement, challenge opens
13 August 2021: submission deadline extended to September 30
21 August 2021: minor changes in text on prediction submission format
30 September 2021: submission deadline extended to October 11
11 October 2021: challenge closed
