Predict the effect of missense mutations on PTEN and TPMT protein stability
Challenge: PTEN and TPMT
Dataset availability: registered users only
Last updated: 1 December 2017
This challenge is closed.
Make sure you understand our Data Use Agreement and Anonymity Policy
Summary
The gene p10 encodes for PTEN (Phosphatase and TEnsin Homolog), an important secondary messenger molecule promoting cell growth and survival through signaling cascades including those controlled by AKT and mTOR. Thiopurine S-methyl transferase (TPMT) is a key enzyme involved in the metabolism of thiopurine drugs and functions by catalyzing the S-methylation of aromatic and heterocyclic sulfhydryl groups. A library of thousands of PTEN and TPMT mutations was assessed to measure the stability of the variant protein using a multiplexed variant stability profiling (VSP) assay, which detects the presence of EGFP fused to the mutated PTEN and TPMT protein respectively. The stability of the variant protein dictates the abundance of the fusion protein and thus the EGFP level of the cell. The challenge is to predict the effect of each variant on TPMT and/or PTEN protein stability.
Background
Deep mutational scans, in which the functional consequences of all possible single nucleotide variants are queried simultaneously (Fowler & Fields, 2014), offer a potential solution to investigate whether a variant is benign or deleterious. However, there is a key problem: developing a bespoke assay for each protein is impractical. Therefore, the Fowler lab and collaborators sought to measure a protein property that could be both informative of variant effect and generalizable to many proteins. Though proteins have a vast range of structures and functions, most proteins share a key requirement: they must be stable enough to perform their role in the cell. Mutations that interfere with thermodynamic stability or folding often cause accelerated turnover and lowered steady-state abundance in cells. Consequently, stability-related reduced protein abundance is a major cause of loss-of-function in monogenic disease (Yue et al., 2005). Loss of tumor suppression activity by destabilizing mutations can lead to cancer, and loss of the enzymes that metabolize drugs can alter drug response.
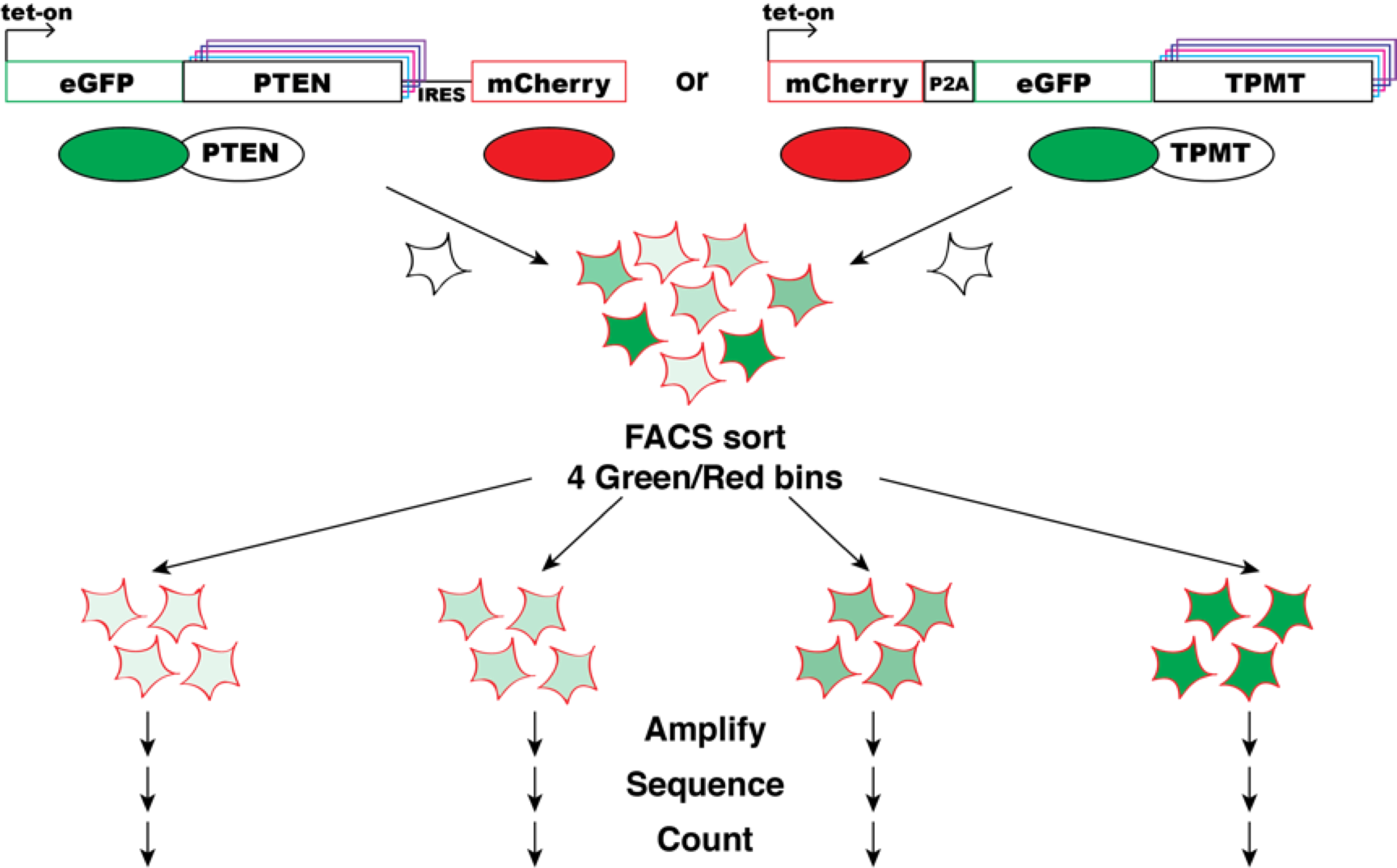
Experiment
A multiplexed variant stability profiling (VSP) assay was developed to measure the steady-state abundance of missense variants of a given protein in human cells. The VSP assay exploits a fluorescent reporter system to measure steady-state abundance of missense protein variants (see figure above). Here, each cell expresses a protein variant fused to EGFP. The stability of the variant protein dictates the abundance of the fusion protein and thus the EGFP level of the cell. As a reporter of transcriptional abundance (Yen et al., 2008), mCherry is either co-transcriptionally or co-translationally expressed from the same construct. Cells are flow sorted into bins according to their EGFP/mCherry ratio, and deep sequencing is used to quantify each variant’s frequency in each bin. Finally, a stability score is calculated based on binwise frequency.
The VSP assay was used to measure the steady state abundance in parallel of thousands of protein variants of Phosphatase and tensin homolog (PTEN) and thiopurine methyltransferase or thiopurine S-methyltransferase (TPMT) protein variants in parallel. Barcoded, site saturation missense libraries of PTEN and TPMT (Jain & Varadarajan, 2014; Starita et al., 2015) were separately recombined into engineered landing pad cells. The EGFP/mCherry ratios of cells harboring each library spanned the range previously characterized for WT and known destabilized variants. Each library was then flow sorted into EGFP/mCherry bins ranging from a low ratio (unstable) to high ratio (stable) variants. Genomic DNA was purified from each bin, and then barcodes were sequenced and tallied. A variant stability score was computed, with 0 meaning unstable, 1 meaning wild-type stability and >1 meaning more stable than wild type.
Variant stability scores correlated well between experimental replicates. Scores and confidence intervals were computed from replicate data for 3,736 PTEN missense variants, and 2,924 TPMT missense variants (60% of 4,900 possible variants).
PTEN (Phosphatase and TEnsin Homolog) is a ubiquitously expressed 403-amino acid protein (Uniprot ID P60484) that dephosphorylates phosphatidylinositol (3,4,5)-triphosphate (PIP3), an important secondary messenger molecule promoting cell growth and survival through signaling cascades including those controlled by AKT and mTOR (Song et al., 2012). Its important regulatory roles in pro-oncogenic processes results in high rates of PTEN missense mutation in diverse cancers including glioma, endometrial cancer, and melanoma. Germline variation in PTEN results in a collection of developmental abnormalities grouped as PTEN Hamartoma Tumor Syndromes (PHTS) (Eng, 2003), and is also associated with autism (Butler et al., 2005).
Thiopurine S-methyl transferase (TPMT) is a key enzyme involved in the metabolism of thiopurine drugs and functions by catalysing the S-methylation of aromatic and heterocyclic sulfhydryl groups (Coelho et al., 2016). TPMT (UniProt ID P51580) is a single domain protein that transfers a S-methyl group from its co-factor S-adenylmethionine to 6-mercaptopurine. The methylated product inhibits de novo purine synthesis leading to cell death. 6-mercaptopurine has been used as a chemotherapeutic agent for Acute-Lymphoblastic Leukemia (ALL) for decades and azathioprine which is converted to 6-mercaptopurine is used to treat autoimmune diseases and to prevent organ rejection after transplant. Overdose with thiopurines leads to treatment interruptions that cause poorer health outcomes and in some cases a life-threatening myelosuppression and hepatotoxicity (Relling et al., 2006).
Prediction challenge
Participants are asked to submit predictions on the effect of each variant on TPMT and PTEN protein stability. The submitted prediction should be a numeric value between 0 (unstable) and 1 (wild-type stability), or >1 (stability is greater than wild-type). Each predicted protein stability must include a standard deviation. Optionally, a comment on the basis of the prediction may be given. The predictions will be assessed against the numeric values of the empirical measurement for each mutation in the assay.
Prediction submission format
The prediction submission is a tab-delimited text file for PTEN and TPMT. Predictors can submit predictions for one or both proteins. Organizers provide a template file, which must be used for submission. In addition, a validation script is provided, and predictors must check the correctness of the format before submitting their predictions. In the submitted file, each row includes the following columns:
- Gene symbol
- Variant
- Prediction - Prediction of relative PTEN and TPMT protein stability - 0 (unstable), 1 (wild-type), or >1 if the predicted stability is greater than wild-type.
- SD of prediction
- Comment - optional brief comment on the basis of the prediction in column 3
In the template file, cells in columns 3-5 are marked with a "*". Submit your predictions by replacing the "*" with your value. No empty cells are allowed in the submission. If you are not confident in a prediction for an individual, enter a suitably large standard deviation for the prediction. Optionally, enter brief comments indicating the basis of the predictions; otherwise, leave the "*" in these cells. Please make sure you follow the submission guidelines strictly.
In addition, your submission must include a detailed description of the method used to make the predictions, similar to the style of the Methods section in a scientific article. This information will be submitted as a separate file.
To submit predictions, you need to create or be part of a CAGI User group. Submit your predictions by accessing the link "All submission forms" from the front page of your group. For more details, please read the FAQ page.
Download dataset: This dataset file is available only to registered users. Please log in to access the file.
Dataset provided by
Kenneth Matreyek (see picture, right), Lea Starita (middle) and Doug Fowler (left) from University of Washington.

References
Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet (2005) 42(4):318-321. PubMed
Coelho T, et al. Genes implicated in thiopurine-induced toxicity: comparing TPMT enzyme activity with clinical phenotype and exome data in a paediatric IBD cohort. Sci Rep (2016) 6:34658. PubMed
Eng C. PTEN: one gene, many syndromes. Hum Mutat (2003) 22(3):183-198. PubMed
Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods (2014) 11(8):801-807. PubMed
Jain PC, Varadarajan R. A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library. Anal Biochem (2014) 449:90-98. PubMed
Relling MV, et al. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood (2006) 107(2):843-844. PubMed
Song MS, et al. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol (2012) 13(5):283-296. PubMed
Starita LM, et al. Massively parallel functional analysis of BRCA1 RING domain variants. Genetics (2015) 200(2): 412-422. PubMed
Yen HC, et al. Global protein stability profiling in mammalian cells. Science (2008) 322(5903):918-923. PubMed
Yue P, et al. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol (2005) 353(2):459-473. PubMed
Revision history
21 October 2017: Initial release
30 October 2017: Closing date updates
24 September 2018: Dataset availability added
