Identify the splicing impact of disease-causing mutations in TP53
Dataset description: public
Exome sequence data: registered users only, limited by CAGI Data Use Agreement
This challenge closed on 25 April 2013.
Splicing Challenge answer key (95 KB, doc): registered users only, limited by CAGI Data Use Agreement
Assessor summary (688 KB, zip): registered users only, limited by CAGI Data Use Agreement
Slides from the CAGI conference: registered users only, limited by CAGI Data Use Agreement
Jeremy Sanford: Assessment (6.6 MB, remixable ppt)
Sean Mooney: Predictor Talk (460 KB, remixable ppt)
Adam Frankish: Predictor Talk (328 kB, remixable ppt)
Predictions (88.5 KB, zip): registered users only, limited by CAGI Data Use Agreement
Background
Accurate precursor mRNA (pre-mRNA) splicing is required for the expression of protein coding genes from the human genome. In this process, intervening sequences (introns) are removed from pre-mRNA and coding/regulatory sequences (exons) are ligated together generating a mature mRNA [1]. Exonic sequences are densely packed regulatory elements such as splicing enhancers and splicing silencers [2,3].
The function of exonic splicing regulatory elements can be undermined by DNA sequence variation and in some cases can contribute to pathogenesis. Thousands of disease-causing mutations disrupt exonic splicing regulatory elements [4,5]. These data suggest that >25 percent of missense mutations may impact pre-mRNA splicing rather than mRNA translation [4,5].
Predicting the functional impact of DNA sequence variants is an important problem. The solution, at least in part, may be found through a mechanistic understanding of how exon sequences contribute to pre-mRNA splicing. Although not fully understood, exonic splicing enhancers are widely believed to promote exon inclusion through a process called exon definition [6,7]. Exon splicing enhancers are also implicated in exon pairing and regulating the inclusion of adjacent exons [8-13].
Using minigene constructs derived from a fragment of the TP53 gene, we have experimentally determined if each mutation influences splicing fidelity in HEK293T cells. We hope that CAGI participants will be able to predict the outcome of our experiments. A long-term goal will be the computational prioritization of disease-causing mutations prior to experimental validation. This contribution is expected to have major impacts in understanding the pathogenic basis of disease-causing mutations.
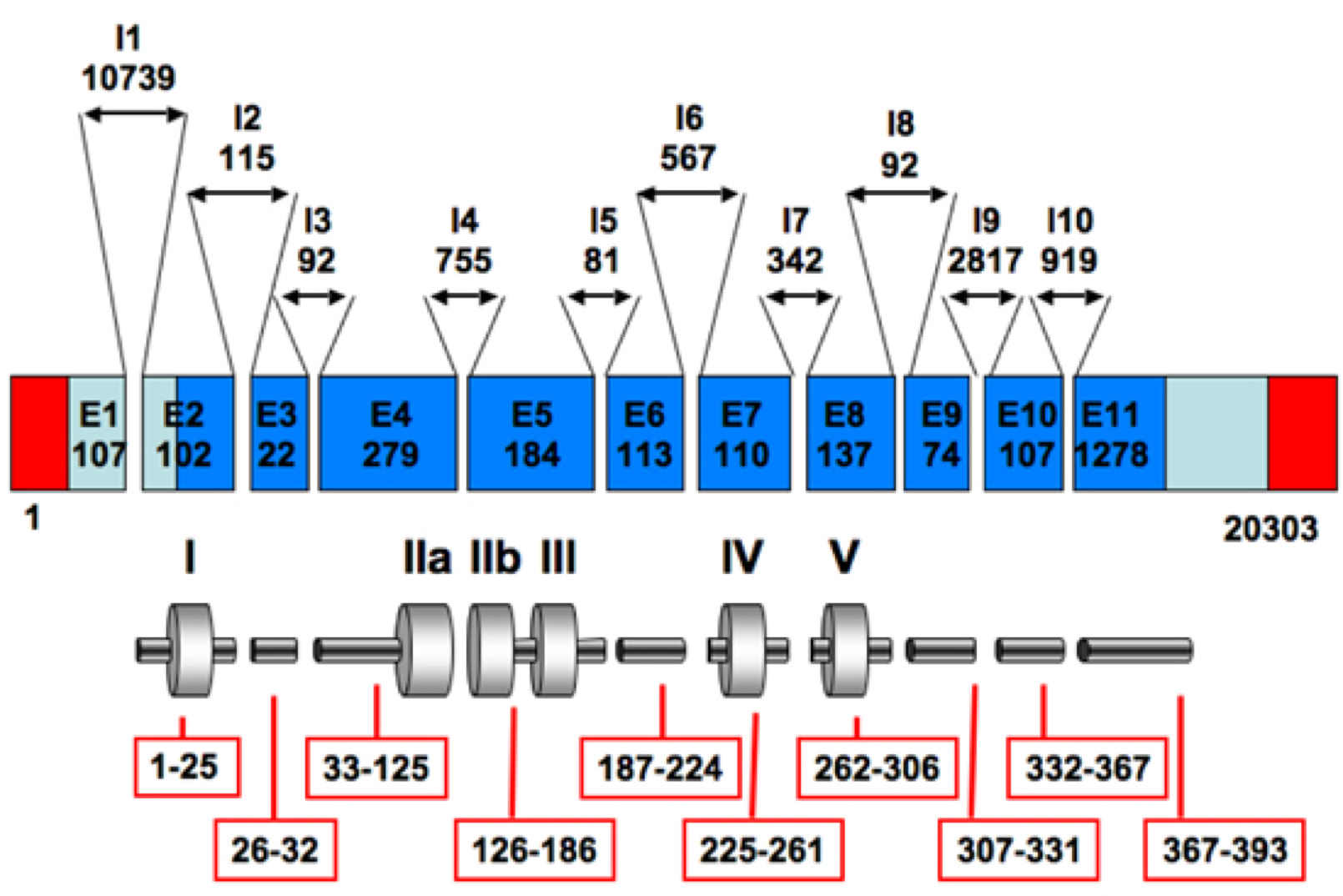
The structure of the TP53 gene is given below, with the translated exons in the blue numbered boxes (from http://p53.free.fr/p53_info/p53_gene.html)
Prediction challenge
With the provided data determine
- which disease-causing mutations in the TP53 gene induce aberrant pre-mRNA splicing and
- which exons (1-11) are defectively spliced.
Dataset: The dataset file is only available for registered users, please log in to access the file.
Prediction submission format
A tab-delimited text file for prediction submissions is provided. There are two header rows: the first designates what is being predicted; the second designates probabilities (P) and standard deviations (SD) for each mutation. Following the two header rows are three rows corresponding to the three mutations presented above, as listed in the first column.
The second column, labeled "Splicing Altered?" contains cells for the probability P (0 – 1) that the splicing is altered for each mutation. The next column holds the standard deviation (SD), defining the confidence of the prediction for that mutation. High SD means low confidence, while small SD means that the predictor is confident about the submitted prediction.
After the Splicing Altered columns, the next 22 columns correspond to sequential exons between 1 and 11, alternating between the probability P (0-1) that that particular exon is altered for each mutation, and the standard deviation (SD), defining the confidence of the prediction for that mutation.
Here is a summary of the column designations in the template.
Mutation Splicing Altered? In Exon1? ...
P SD P SD P SD
1 * * * * * *
2 * * * * * *
3 * * * * * *
In the template file, all blank cells are marked with an "*". Submit your predictions by replacing the "*" with your value. No empty cells are allowed in the submission; if you cannot submit predictions for a variant, please leave the sign "*" in these cells. Please make sure you follow these submission guidelines strictly.
In addition, a validation script is provided, and predictors should check the correctness of the format before submitting their predictions.
Download submission template
Download Validation script (not available).
Dataset provided by

Linnea Jannson, Tim Sterne-Weiler, and Jeremy Sanford (picture).
References
- Solis, A.S., N. Shariat, and J.G. Patton. 2008. Splicing fidelity, enhancers, and disease. Front Biosci 13: 1926-1942.
- Fairbrother, W.G., D. Holste, C.B. Burge, and P.A. Sharp. 2004. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol 2: E268.
- Ke, S., X.H. Zhang, and L.A. Chasin. 2008. Positive selection acting on splicing motifs reflects compensatory evolution. Genome Res 18: 533-543.
- Sterne-Weiler, T., J. Howard, M. Mort, D.N. Cooper, and J.R. Sanford. 2011. Loss of exon identity is a common mechanism of human inherited disease. Genome Res 21: 1563-1571.
- Sterne-Weiler, T., J. Howard, M. Mort, D.N. Cooper, and J.R. Sanford. 2011. Loss of exon identity is a common mechanism of human inherited disease. Genome Res 21: 1563-1571.
- Lim, K.H., L. Ferraris, M.E. Filloux, B.J. Raphael, and W.G. Fairbrother. 2011. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A 108: 11093-11098.
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J Biol Chem 270: 2411-2414.
- Schaal, T.D. and T. Maniatis. 1999. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol 19: 261-273.
- Schneider, M., C.L. Will, M. Anokhina, J. Tazi, H. Urlaub, and R. Luhrmann. 2010. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol Cell 38: 223-235.
- Sanford, J.R., P. Coutinho, J.A. Hackett, X. Wang, W. Ranahan, and J.F. Caceres. 2008. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS ONE 3: e3369.
- Han, J., J.H. Ding, C.W. Byeon, J.H. Kim, K.J. Hertel, S. Jeong, and X.D. Fu. 2011. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol 31: 793-802.
- Sanford, J.R., X. Wang, M. Mort, N. Vanduyn, D.N. Cooper, S.D. Mooney, H.J. Edenberg, and Y. Liu. 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381-394.
- Cramer, P., J.F. Caceres, D. Cazalla, S. Kadener, A.F. Muro, F.E. Baralle, and A.R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251-258.
- Ghigna, C., S. Giordano, H. Shen, F. Benvenuto, F. Castiglioni, P.M. Comoglio, M.R. Green, S. Riva, and G. Biamonti. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20: 881-890.
Assessment
This challenge is being assessed by Jeremy Sanford, University of California, Santa Cruz.
